南湖新闻网讯(通讯员 黄宇虹)近日,我校作物遗传改良全国重点实验室、湖北洪山实验室小麦遗传改良创新团队苏汉东教授课题组联合中国科学院遗传发育所的合作研究成果以“Distinct evolutionary trajectories of subgenomic centromeres in polyploid wheat”为题在Genome Biology发表。该研究通过对普通小麦及其二倍体、四倍体祖先进行系统性的比较基因组学分析,绘制出了精细而动态的小麦着丝粒演化图谱,发现在异源六倍体小麦中,来自不同祖先AA、BB和DD亚基因组的着丝粒,在多倍化后走上了截然不同的演化道路。
着丝粒是确保细胞分裂时染色体能够被精准、均等地分配到子细胞中的核心结构。尽管其功能在所有真核生物中高度保守,着丝粒DNA序列却是基因组中进化最迅速的部分之一,这一现象被称为“着丝粒悖论”。在多倍体植物中,这一悖论显得尤为复杂。多倍体植物的细胞核内,汇集了来自不同祖先物种的染色体组,它们的着丝粒被迫在同一个细胞核环境中共同工作和演化。这些来自不同“家乡”的着丝粒是如何在多倍化这一剧烈的基因组事件后相互适应、协同进化的?它们的演化遵循着怎样的规律?这些问题一直是基因组进化生物学领域的谜团。
普通小麦拥有清晰的、阶梯式的多倍化历史,是研究这一科学问题的绝佳模型。本研究通过整合多物种、多倍性水平的基因组和表观基因组数据,系统揭示了异源多倍体小麦中不同亚基因组着丝粒的演化“命运”大相径庭(如图)。研究发现,小麦着丝粒的演化主要由着丝粒特异性逆转录转座子(CRWs)驱动,但不同亚基因组采取了不同的演化策略:AA亚基因组通过新CRW家族的不断“入侵”和对旧序列的“替换”来进行动态重塑;而DD亚基因组则通过向侧翼区域的“扩张”来适应新的核环境,且这一过程是渐进的。这项工作不仅为理解复杂基因组中着丝粒的演化和维持机制提供了见解,也展现了多倍化后基因组内部不同组分如何通过差异化的路径实现协同演化。这些发现为利用远缘杂交进行小麦遗传改良提供了理论基础,有助于更好地理解和预测杂交后代基因组的稳定性和演化动态。
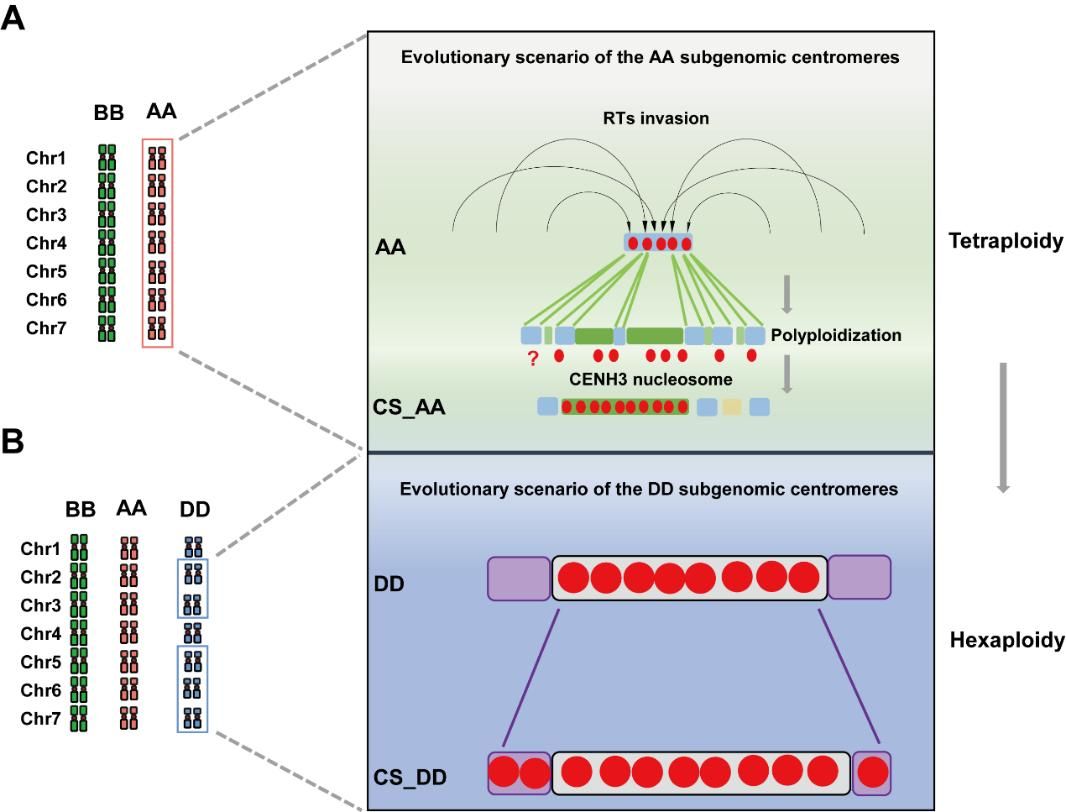
多倍体小麦中亚基因组着丝粒的独特演化模式图
华中农业大学作物遗传改良全国重点实验室博士后黄宇虹、中国科学院遗传发育所刘阳副研究员与刘畅博士为论文共同第一作者,华中农业大学作物遗传改良全国重点实验室、湖北洪山实验室苏汉东教授与中国科学院遗传发育所韩方普研究员为论文共同通讯作者。该研究得到国家重点研发计划和国家自然科学基金等项目资助。
审核人:苏汉东
论文链接:https://genomebiology.biomedcentral.com/articles/10.1186/s13059-025-03759-4
英文摘要:
Background:Centromeres are crucial for precise chromosome segregation and maintaining genome stability during cell division. However, their evolutionary dynamics, particularly in polyploid organisms with complex genomic architectures, remain largely enigmatic. Allopolyploid wheat, with its well-defined hierarchical ploidy series and recent polyploidization history, serves as an excellent model to explore centromere evolution.
Results:In this study, we perform a systematic comparative analysis of centromeres in common wheat and its corresponding ancestral species, utilizing the latest comprehensive reference genome assembly available. Our findings reveal that wheat centromeres predominantly consist of five types of centromeric-specific retrotransposon elements (CRWs), with CRW1 and CRW2 being the most prevalent. We identify distinct evolutionary trajectories in the functional centromeres of each subgenome, characterized by variations in copy number, insertion age, and CRW composition. By utilizing CENH3-ChIP data across various ploidy levels, we uncover a series of CRW invasion events that have shaped the evolution of AA subgenome centromeres. Conversely, the evolutionary process of the DD subgenome centromeres involves their expansion from diploid to hexaploid wheat, facilitating adaptation to a larger genomic context. Integration of complete einkorn centromere assemblies and Aegilops tauschii pan-genomes further revealed subgenome-specific centromere evolutionary trajectories. By inclusion of synthetic hexaploid from S2-S3 generations, alongside 2x/6 × natural accessions, we demonstrate that DD subgenome centromere expansion represents a gradual evolutionary process rather than an immediate response to polyploidization.
Conclusions:Our study provides a comprehensive landscape of centromere adaptation, evolution, and maturation, along with insights into how retrotransposon invasions drive centromere evolution in polyploid wheat.